Estimation of parameters¶
In this tutorial, we show an detailed example of how you can use a pre-trained neural network to estimate biomarkers from input signals. We use the network that we trained to obtain the results presented in our paper.
[1]:
import sys
depth = '../'
for i in range(5):
sys.path.append(depth)
depth += '../'
import os
from os.path import isfile, join
import importlib
import numpy as np
import torch
import scipy as sc
from scipy.io import loadmat
import pickle
import MRF
from MRF.Training_parameters import *
from MRF.BaseModel import *
from MRF.Projection import *
from MRF.models import *
from MRF.Offline import Network, Data_class, Performances
import matplotlib.pyplot as plt
root = '../../../../'
1) Description of the processed signals¶
In order to preserve anonymity of MRI volunteers, we do not provide the data corresponding to the reconstructed images obtained from a full scan. We will be focused on a specific slice and show how we can use our pre-trained networks to get results. The data corresponding to this specific slice can be found in the folder paper_data.
[2]:
test_slice = sc.io.loadmat(root+'paper_data/invivo_test_slice.mat')['x']
2) Parameters estimation¶
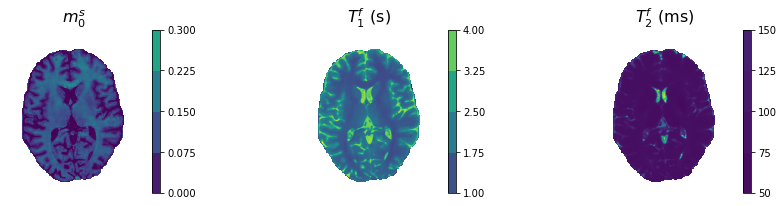
We use our pre-trained neural network to estimate the three parameters \(m_0^s\), \(T_1^f\) and \(T_2^f\).
We define a directory to save results.
[3]:
save_root='/NN_recon/'
if not os.path.exists(save_root):
os.makedirs(save_root)
We give the list of the names of the trained networks that we want to use to process data.
[4]:
names = [ 'CRB-paper']
We give the list of paths where we saved the input signals that we want to process using the above mentioned pre-trained networks.
[5]:
data_files = ['paper_data/invivo_test_slice.mat']
[6]:
with open(root+'settings_files_offline/settings_'+name+'.pkl', 'rb') as f:
settings = pickle.load(f)
print(settings)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
<ipython-input-6-9ceb51fc5ad2> in <module>
----> 1 with open(root+'settings_files_offline/settings_'+name+'.pkl', 'rb') as f:
2 settings = pickle.load(f)
3 print(settings)
NameError: name 'name' is not defined
We compute the estimate of the parameters and we save the results.
[7]:
t_1 = time.time()
for name, data_file in zip(names, data_files):
filepath = os.path.join(root, data_file)
with open(root+'settings_files_offline/settings_'+name+'.pkl', 'rb') as f:
settings = pickle.load(f)
##################
# Neural Network Architecture #
##################
model = importlib.import_module('MRF.models.' + settings['name_model'])
settings['namepca'] = root+'/paper_data/basis_for_compress.mat'
##################
# Loading the pre-trained network #
##################
net = torch.load(join(root,'save_networks_offline/network_'+settings['name_model']),map_location='cpu')
training_parameters = Training_parameters(settings['batch_size'], 1, settings['nb_epochs'], settings['params'], settings['normalization'], settings['complex'])
projection = Projection(settings['start_by_projection'], settings['dimension_projection'], settings['initialization'], settings['normalization'], settings['namepca'], settings['complex'])
data_class = Data_class(training_parameters, settings['noise_type'], settings['noise_level'],
settings['minPD'], settings['maxPD'], settings['nb_files'], settings['path_files'])
validation_settings = {'validation': settings['validation'],'small_validation_size': settings['small_validation_size'], 'validation_size': settings['validation_size']}
netw = model.model(projection=projection,nb_params=len(settings['params']))
device = torch.device('cpu')
try:
netw.load_state_dict(net['NN'])
except:
projection = Projection(settings['start_by_projection'], settings['dimension_projection'], 'Fixlayer', settings['normalization'], settings['namepca'], settings['complex'], ghost=True)
netw = model.model(projection=projection,nb_params=len(settings['params']))
netw.load_state_dict(net['NN'])
netw.eval()
##################
# Importing data #
##################
import h5py
import numpy as np
try:
from scipy.io import loadmat
arrays = loadmat(filepath)
fingers = arrays['x']
fingers = fingers.T
except:
arrays = {}
f = h5py.File(filepath, 'r')
for k, v in f.items():
arrays[k] = np.array(v)
fingers = arrays['x']
t_2 = time.time()
##################
# Preparing the neural network by removing the potential first projection layer #
##################
projection.initialization = 'Fixlayer'
netwproj = model.model(projection=projection,nb_params=len(settings['params']))
device = torch.device('cpu')
dico = net['NN']
try:
if net['complex']:
del dico['fc1_real.weight']
del dico['fc1_imag.weight']
else:
del dico['fc1.weight']
except:
pass
mrfshape = fingers.shape
netwproj.load_state_dict(dico, strict=False)
netwproj.eval()
##################
# Estimating the biophysical parameters #
##################
with torch.no_grad():
if len(mrfshape)<=3:
mrfshape = np.hstack((mrfshape,[1]))
fings = fingers.reshape((-1, mrfshape[1],mrfshape[2] * mrfshape[3]))
sequence_to_stack = []
for i in range(mrfshape[1]):
fings_tmp = fings[:,i,:].T
params_tmp = netwproj(torch.tensor(fings_tmp, dtype=torch.float))
params_tmp = np.array(params_tmp)
for ii, para in enumerate(settings['params']):
if settings['loss'][para] == 'MSE-Log':
params_tmp[:, ii] = 10 ** params_tmp[:, ii]
params_tmp = params_tmp.reshape((mrfshape[2],mrfshape[3],len(settings['params'])))
sequence_to_stack.append(params_tmp)
params = np.stack(sequence_to_stack,axis=0)
##################
# Saving results and displaying processing time #
##################
processing_time = time.time() - t_2
params = np.moveaxis(params, [0, 2], [2, 0])
total_time = time.time() - t_1
print(os.path.join(save_root, 'qM_'+data_file[2:-4]+ '_'+name+'.mat'))
print('done results')
print('processing_time')
print(processing_time)
print('total_time')
print(total_time)
/NN_recon/qM_per_data/invivo_test_slice_CRB-paper.mat
done results
processing_time
3.6539628505706787
total_time
3.903989315032959
[8]:
fig, ax = plt.subplots(len(net['params'])//3,3, figsize=(15,3))
ranges = [[0,0.3],[1,4],[50,150]]
scaling = [1,1,1e3]
units = ['',' (s)',' (ms)']
for i, ind in enumerate(net['params']):
shw = ax[i].imshow(scaling[i]*params[0,:,:,i].T)
ax[i].set_title(paramtolatexname[ind]+units[i], size=16)
plt.colorbar(shw, ax=ax[i], boundaries=np.linspace(ranges[i][0],ranges[i][1],5))
ax[i].axis('off')
plt.show()